바이오사이토젠의 유전자 편집 플랫폼은 2009년 회사의 창립 이래, 동물 및 세포모델의 연구개발을 수행하고 있습니다. 유전자 편집 모델은 기초과학연구 분야에서만 큰 영향을 미칠 뿐만 아니라, 항체신약의 연구개발 과정에서 발생하는 중요한 문제들을 해결해주는 역할을 합니다. 완전한 인간 항체가 생산 가능한 RenMab마우스를 사용하여 완전한 인간 서열을 가지면서 동시에 마우스의 체내 면역 시스템 내에서 선별된 항체 분자를 직접 얻을 수 있습니다. 동일한 light chain의 항체를 생산할 수 있는 RenLite 마우스는 이중특이성 항체 생산시의 단백질 발현 과정중의 잘못된 조합 문제를 해결할 수 있습니다. 또한 heavy chain only의 항체를 생산할 수 있는 RenNano마우스를 직접 사용하여 완전한 인간 나노항체를 얻을 수 있습니다.
그 외에도, 일반적인 방법으로 얻어진 항체 분자는 인간에서의 표적만 식별할 수 있으며 마우스에서의 동일한 표적은 식별할 수 없어서, 타겟의 인간화 마우스/세포모델을 사용하여야만 이러한 약물의 안전성 평가, 약효 평가, 작용 기전 연구등을 수행할 수 있습니다. 이러한 응용 방법 이외에도, 유전자 편집 모델의 사용 범위는 엄청나게 광범위합니다. 예시 : 특정 유전자를 수정하여 항원에의 면역 반응을 높일 수 있으며, 유전적 개조를 통하여 질병 모델을 유도하고 제작할 수 있는 등의 응용법이 있습니다.
수년간의 독자적인 기술연구개발을 통하여, 바이오사이토젠은 3가지의 핵심적인 유전자 편집 기술 시스템을 구축하였습니다: (1) 마우스의 배아 간세포에 기반한 ESC/HR기술을 통하여 제작한 세포주는 70회 가까운 계대 배양에도 여전히 완전한 기능 특성을 보여줬으며, 한 염색체 내에서 여러 유전자의 개조에도 성공하였습니다. (2) 기존의 CRISPR/Cas9을 사용한 기술보다 유전자 Knock-in 효율이 최대 20배까지 높은 EGE기술을 보유하고 있습니다. (3) DNA의 단편 길이에 영향을 받지 않고, 최대 3번의 마우스 배아간세포에서의 조작을 통해 1회성의 매우 긴 인간화 유전자 단편을 얻을 수 있는 SUPCE기술이 있습니다. 이 세가지 기술 시스템을 조합하여, 우리는 기존 유전자 기술 플랫폼들에서 만연한 문제들을 극복하여, 고효율, 대규모, 중복 편집, 긴 유전자 단편의 인간화 등을 바탕으로 임의의 길이와 위치에서의 정확한 유전자 편집을 대부분 문제없이 실현할 수 있게 되었습니다.
ESC/HR
ESC/HR (ESC, Embryonic Stem Cell; HR, Homologous Recombination)는 마우스의 배아간세포에서 DNA와 게놈 사이에서의 동원 접합을 이용하는 유전자 편집 기술 중 한가지입니다. 유전자 개조에서의 양성 클론을 마우스의 낭포에 주사하고, 후속의 번식 과정을 통하여 완성된 성체 마우스를 얻어낼 수 있습니다. 바이오사이토젠은 자체적으로 C57BL/6 백그라운드의 간세포 시스템과 배양 체계를 구축하였고, 100회 가까운 계대배양 후에도 완전한 기능성을 유지하고 있는 성체 마우스를 제작할 수 있습니다.
EGE
EGE (Extreme Genome Editing system)는 CRISPR/Cas9기술을 근간으로 하여, 독자적인 연구개발을 거쳐 유전자의 Knock-in 효율을 약 20배 가까이 상승시킨 기술입니다. CRISPR/Cas9기술이 동원접합의 발생 확률을 극대화하고, 유전자 편집 프로세스를 간략화하였지만, 대량의 모델을 제작하기에는 효율이 낮고 비용이 많이 발생하는 문제가 있습니다. EGE 기술을 사용하면 유전자 편집을 더욱 단시간내에 간편하게 할 수 있으며, 거의 어떠한 게놈의 DNA서열이라도 정확하게 편집이 가능합니다. 유전자 편집 효율을 높이면 더욱 많은 장점이 생기게 됩니다. 첫째로, 동물 모델을 제작하는데 있어서, 성공률을 높이고 비용을 절약하며 제작 주기를 단축할 수 있어, 공업화된 대규모의 상업적 응용에 있어서 큰 장점이 있습니다. 그 다음으로는 각기 다른 유전자의 편집의 난이도가 전부 다름을 고려하여, 편집 난이도가 매우 높은 게놈에 대하여, EGE기술은 불가능을 가능으로 바꿀 만큼 높은 효율성을 자랑합니다.
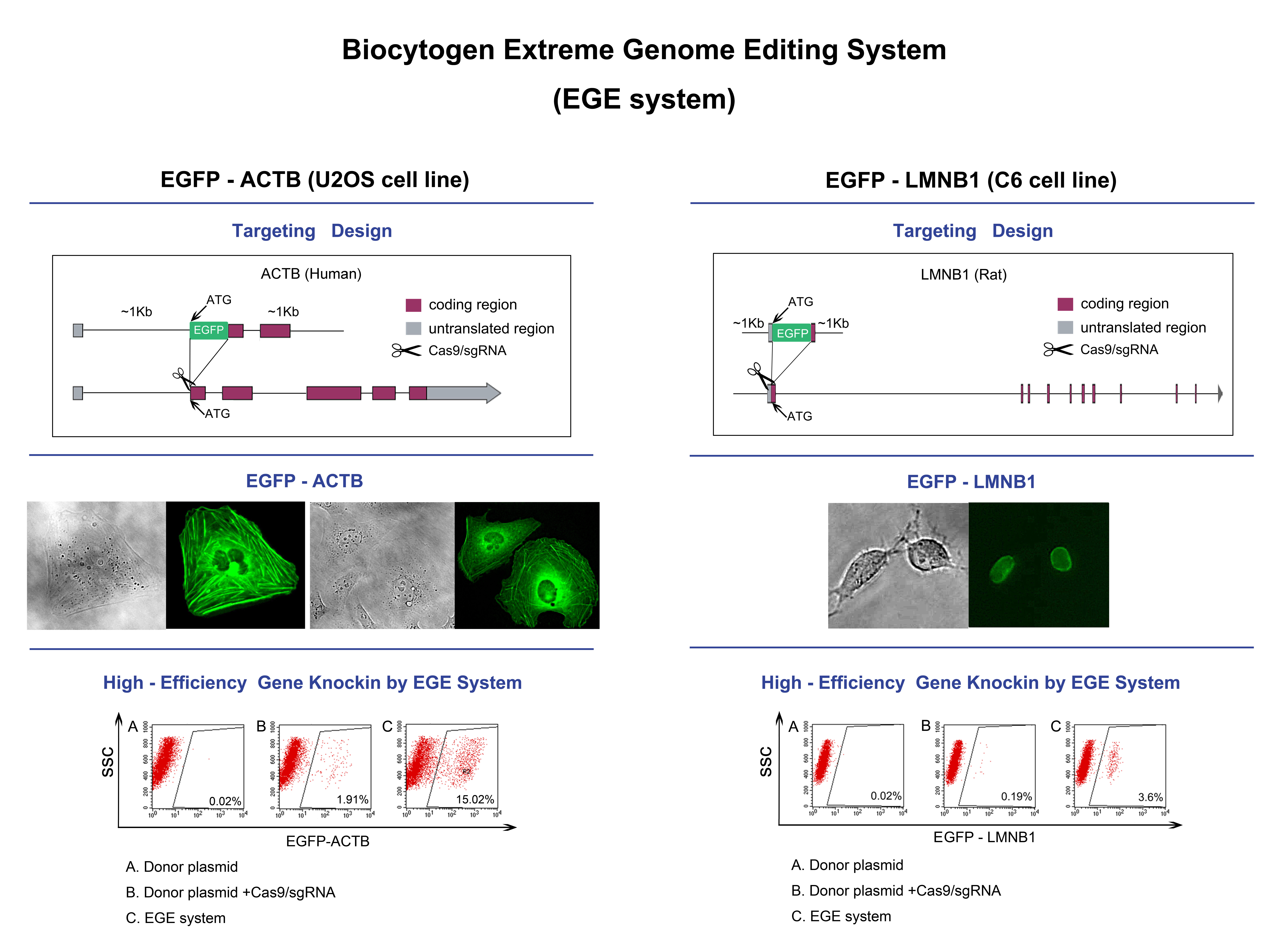
그림1. EGE기술은 CRISPR/Cas9의 유전자 Knock-in 효율을 대폭 상승시켰습니다.
그림 왼쪽의 1번 모델에서 인간의 골육종 세포를 사용하여, 세포의 골격 단백질인 β-Actin의 유전자 ACTB의 개시코돈인 ATG의 뒤쪽에 녹색 형광 단백질인 EGFP를 Knock-in하였으며, 성공적으로 편집된 세포는 EGFP가 융합된 β-Actin을 발현하게 됩니다. U2OS세포는 Cas9 핵산 효소와 ACTB를 타겟하는 sgRNA 플라스미드를 발현하며, 1kb길의의 Homologous arm의 EGFP 제공 플라스미스를 양쪽에 포함하고 있습니다. 플라스미드 감염 후 FACS분석을 통하여, 기본적인 Cas9/sgRNA를 통한 Knock-in 효율은 1.91%, EGE기술을 이용한 Knock-in 효율은 15.02%에 달한다는 것을 확인할 수 있습니다. 사진 오른쪽의 두번째 모델에서는, 랫트의 뇌교종인 C6세포를 선택하여 랫트에서 핵막을 코딩하는 단백질의 유전자인 Lmnb1을 타겟으로 하였으며, EGE기술을 사용하여 기존의 Knock-in 효율인 0.19%를 3.6%로 높였습니다.
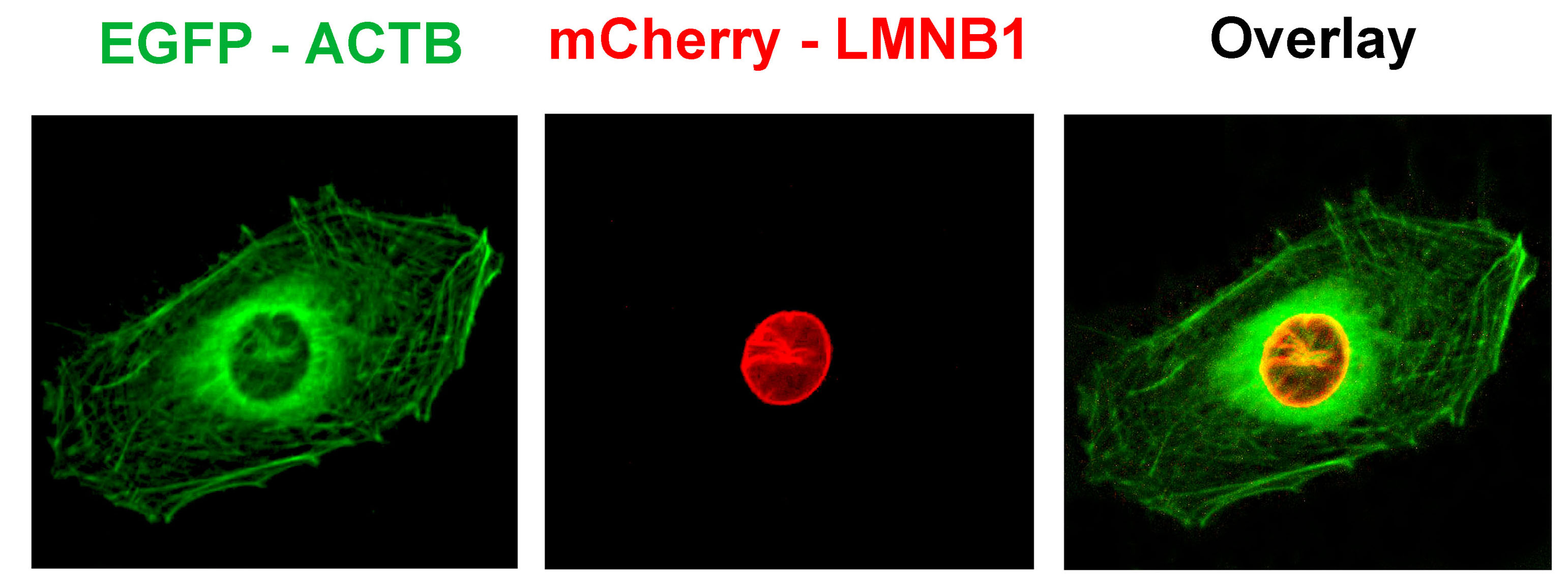
그림2. EGE기술을 통하여 세포에서 동시에 2개의 유전자를 편집하는 것이 가능합니다.
U2OS세포에서 ACTB,LMNB1유전자를 각각 타겟하는 Cas9/sgRNA플라스미드 (ACTB를 표지하는 녹색 형광 단백질 GFP 제공 플라스미드, LMNB1를 표지하는 붉은색 형광 단백질 mCherry 제공 플라스미드) 를 개별적으로 감염시켰습니다. 감염 후의 형광을 현미경으로 관찰한 결과, 붉은색의 핵막단백질과 녹색의 세포 골격 단백질을 관찰할 수 있었습니다.
SUPCE
SUPCE (Size-Unlimited and Precise Chromosome Engineering system) 는 DNA단편의 길이에 제한이 없는 유전자 편집 기술입니다. 기존에 사용된 플라스미드나 BAC(Bacterial Artificial Chromosome) 기술은 긴 단편을 마우스의 배아간세포에 치환하기 위해서는 여러번의 편집이 필요했습니다. 용량의 한계 때문에, 유전자 편집 횟수와 치환가능한 단편의 길이는 정비례합니다. 마우스의 ES세포를 체외에서 장시간 다룰 때 기능을 잃는 일이 매우 빈번하게 발생하여 성체 마우스를 얻지 못하는 일이 자주 발생합니다. SUPCE기술은 이러한 문제를 본질적인 부분에서부터 해결했습니다.
SUPCE기술을 이용하여, 바이오사이토젠은 완전한 인간 항체가 생산 가능한 RenMab의 개발을 성공적으로 끝마쳤으며(그림 1), 그림2의 FISH검사 결과를 통하여 마우스 게놈의 상동 위치에 인간 항체의 heavy chain과 kappa light chain이 성공적으로 치환되었음을 확인할 수 있습니다.
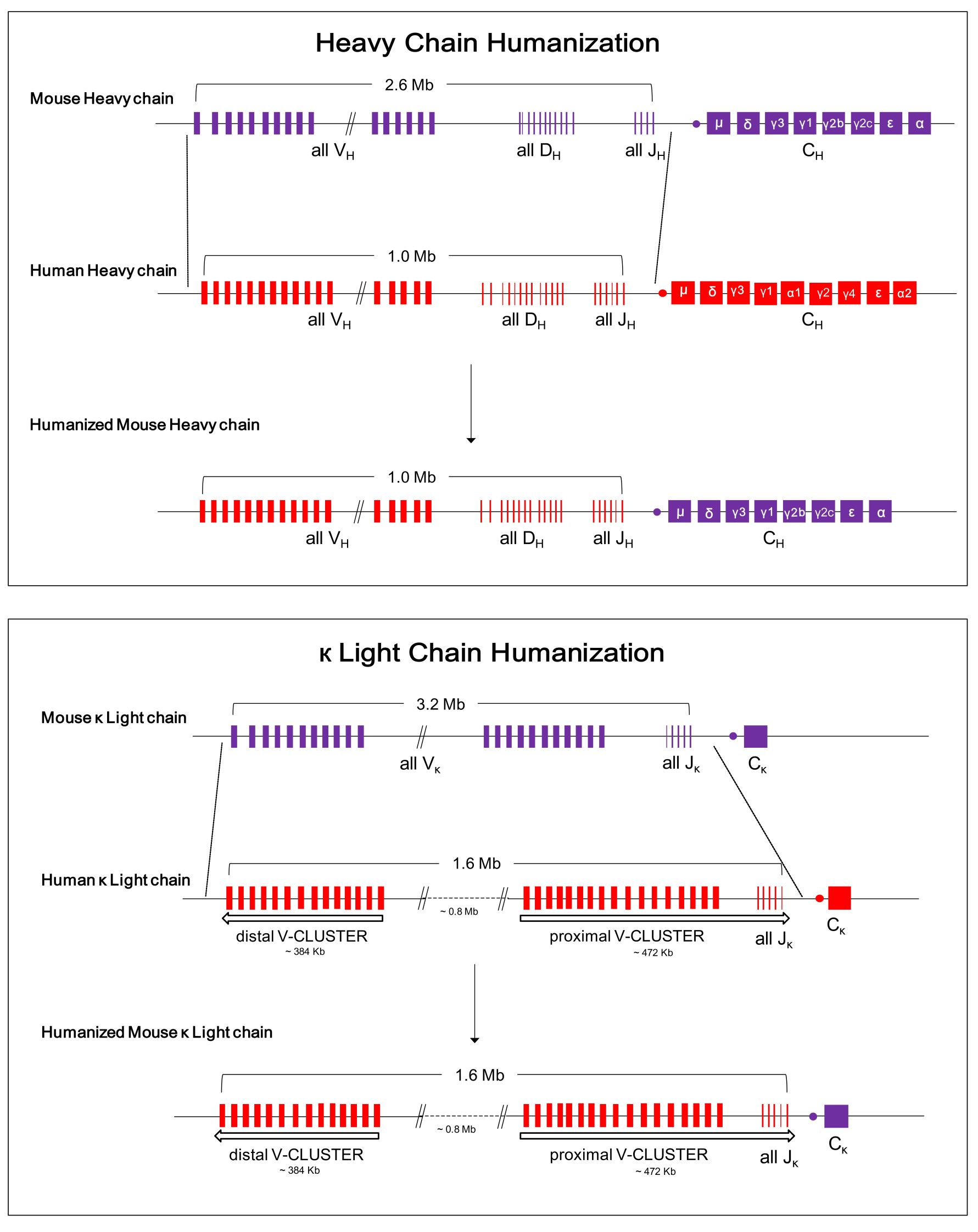
그림 1. RenMab마우스에서의 항체 유전자 인간화 설계 모식도.
RenMab마우스에서, 2.6 Mb길이의 마우스 heavy chain이 1.0 Mb길이의 인간 heavy chain가변영역 서열로 치환되었으며, 3.2 Mb길이의 마우스 kappa light chain의 가변영역은 1.6 Mb길이의 인간의 서열로 대체되었습니다.
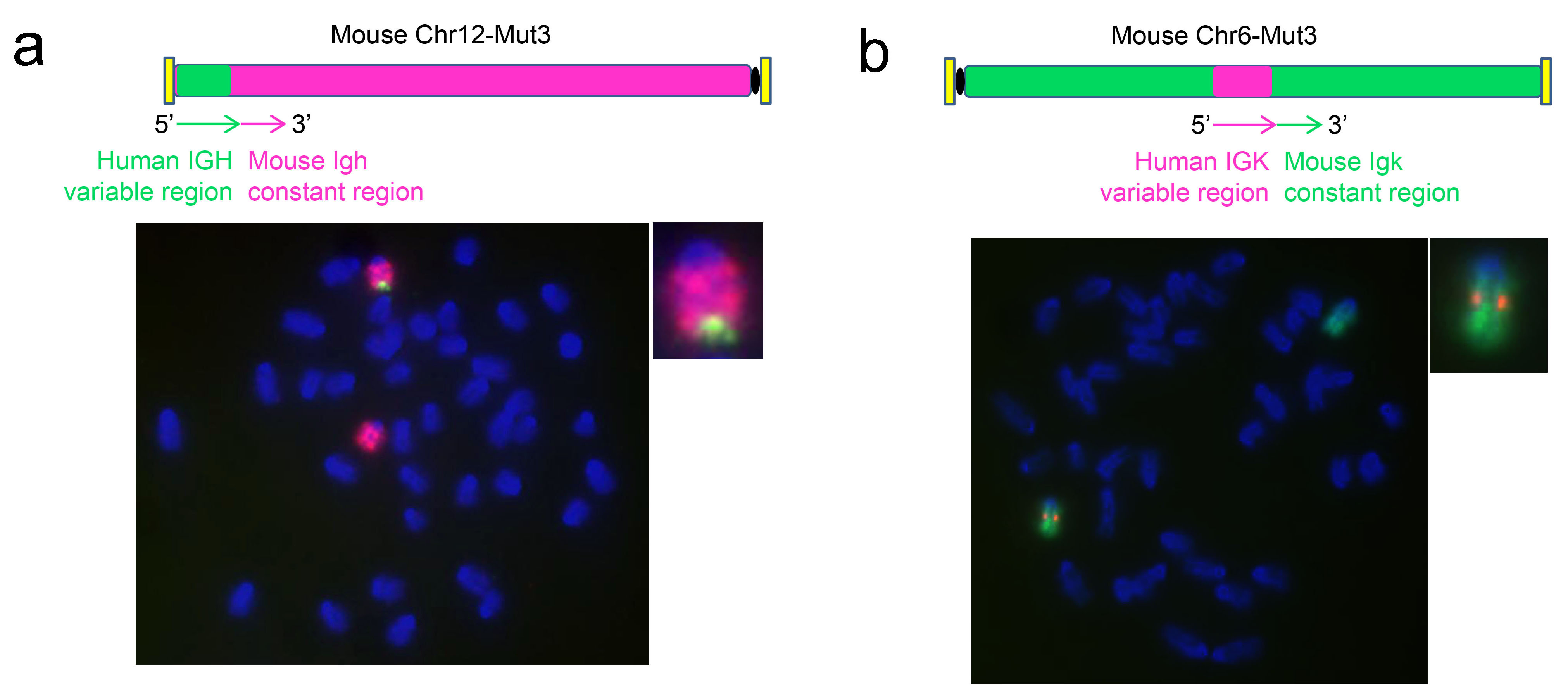
그림2. 항체의 유전자 가변영역의 인간화를 확인하는 FISH검사 결과.
SUPCE기술을 마우스의 배아간세포에 적용하여 마우스의 heavy chain, kappa light chain의 가변영역을 인간 서열로 완전히 치환하였습니다. a. 인간의 heavy chain가변영역 서열 분석 (녹색), 마우스의 Igh유전자가 위치한 12번 염색체의 전체 염색 결과 (홍색), heavy chain인간화 세포 클론 분석 결과. b. 인간의 kappa light chain가변영역 서열 분석 (홍색), 마우스의 Igk 유전자가 위치한 6번 염색체의 전체 염색 결과 (녹색), kappa light chain인간화 세포 클론 분석 결과.
이러한 자주적으로 연구개발한 유전자 편집 기술 플랫폼 이외에도, 자동화되고 대용량의 처리가 가능한 기술 프로세스를
통하여 대량의 모델 준비를 빠르고 안정적으로 끝낼 수 있게 되었습니다. 96채널의 핵산 추출 시스템 여러대를 이용하여 마우스/세포의 유전자 DNA와 플라스미드의 DNA의 취득이 가능하며, 전자동화된 Liquid
handling system을 사용하여 유전자형의 검측의 전자동화를 실현하였습니다. 이러한 플랫폼들을 통하여 매년 1500가지 이상의 모델 개발을 완성해내고 있습니다.