B-hHLA-DRB1*1.1 mice(C)
| Strain Name |
BALB/cCrSIcNifdc-H2-Ea1tm1(HLA-DRA1*1.1-I-Ea)Bcgen H2-Eb1 tm1(HLA-DRB1*1.1-I-Eb)Bcgen H2-Atm1Bcgen H2-Eb2tm1Bcgen/Bcgen |
Common Name | B-hHLA-DRB1*1.1 mice(C) |
| Background | BALB/cCrSIcNifdc | Catalog number |
112754 |
|
Related Genes |
HLA-DRA1; SS1, DRB1, HLA-DRB, HLA-DR1B |
||
|
NCBI Gene ID |
3122, 3123 | ||
- HLA-DRA1 belongs to the HLA class II alpha chain paralogues. HLA-DRB1 belongs to the HLA class II beta chain paralogues. This class II molecule is a heterodimer consisting of an alpha (HLA-DRA) and a beta (HLA-DRB) chain, both anchored in the membrane. It plays a central role in the immune system by presenting peptides derived from extracellular proteins. Class II molecules are expressed in antigen presenting cells (APC: B lymphocytes, dendritic cells, macrophages). The alpha chain is approximately 33-35 kDa and its gene contains 5 exons. The beta chain is approximately 26-28 kDa and its gene contains 6 exons.
- The exons 2-4 of mouse I-Ea1 gene that encode the extracellular and transmembrane domain were replaced by human counterparts in B-hHLA-DRB1*1.1 mice(C). The exons 2-4 of mouse I-Eb1 gene that encode the extracellular and transmembrane domain were replaced by human counterparts in B-hHLA-DRB1*1.1 mice(C). The mouse I-Aa, I-Ab and I-Eb2 were knocked out in B-hHLA-DRB1*1.1 mice(C).
- Human HLA-DR was exclusively detectable in homozygous B-hHLA-DRB1*1.1 mice(C), but not in wild-type BALB/cCrSIcNifdc mice.
- B-hHLA-DRB1*1.1 mice(C) provide a powerful preclinical model for in vivo evaluation of vaccines.
- Application: For example, this product is used for pharmacodynamics and safety evaluation of vaccines for cancers.
Gene targeting strategy for B-hHLA-DRB1*1.1 mice(C). The exons 2-4 of mouse I-Ea1 gene that encode the extracellular and transmembrane domain were replaced by human counterparts in B-hHLA-DRB1*1.1 mice(C). The exons 2-4 of mouse I-Eb1 gene that encode the extracellular and transmembrane domain were replaced by human counterparts in B-hHLA-DRB1*1.1 mice(C). The mouse I-Aa, I-Ab and I-Eb2 were knocked out in B-hHLA-DRB1*1.1 mice(C).
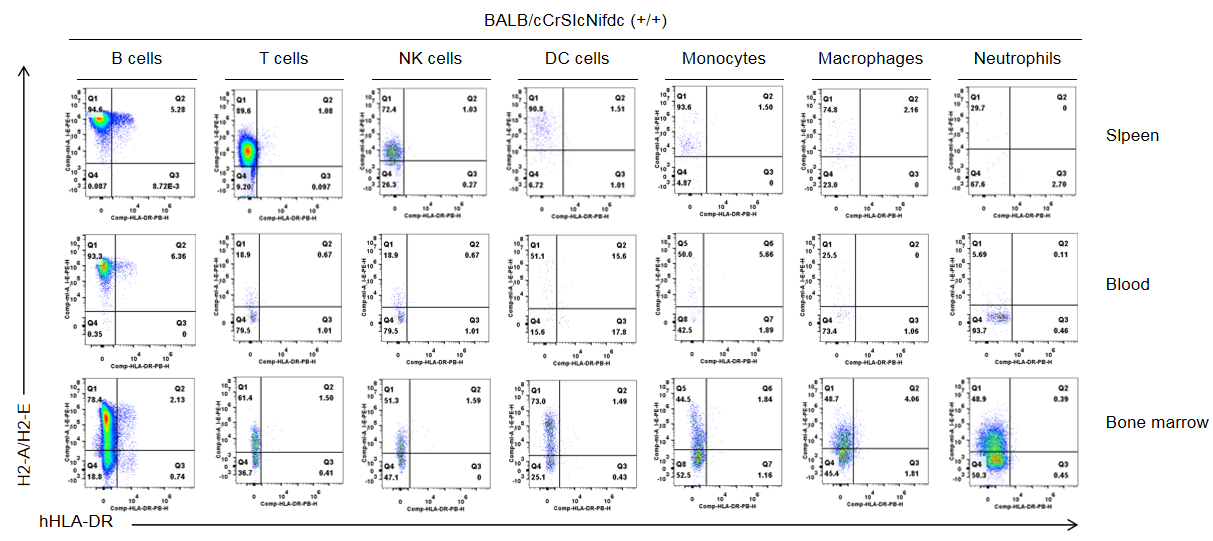
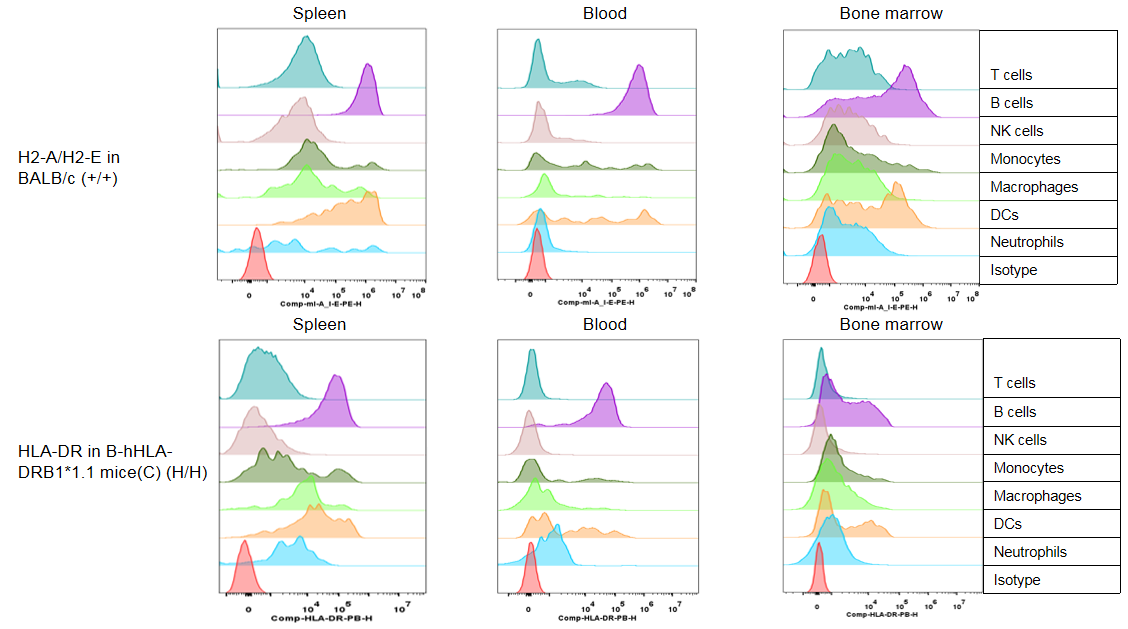
Strain specific HLA-DR expression analysis in wild type BALB/cCrSIcNifdc mice by flow cytometry. Splenocytes, blood cells and bone marrow cells were collected from wild-type BALB/cCrSIcNifdc mice (+/+), and analyzed by flow cytometry with species-specific anti-mouse H2-A/H2-E antibody(Biolegend, 107607), and anti-HLA-DR antibody(Biolegend, 307624). Mouse H2-A/H2-E was only detectable in immune cells of wild-type mice.
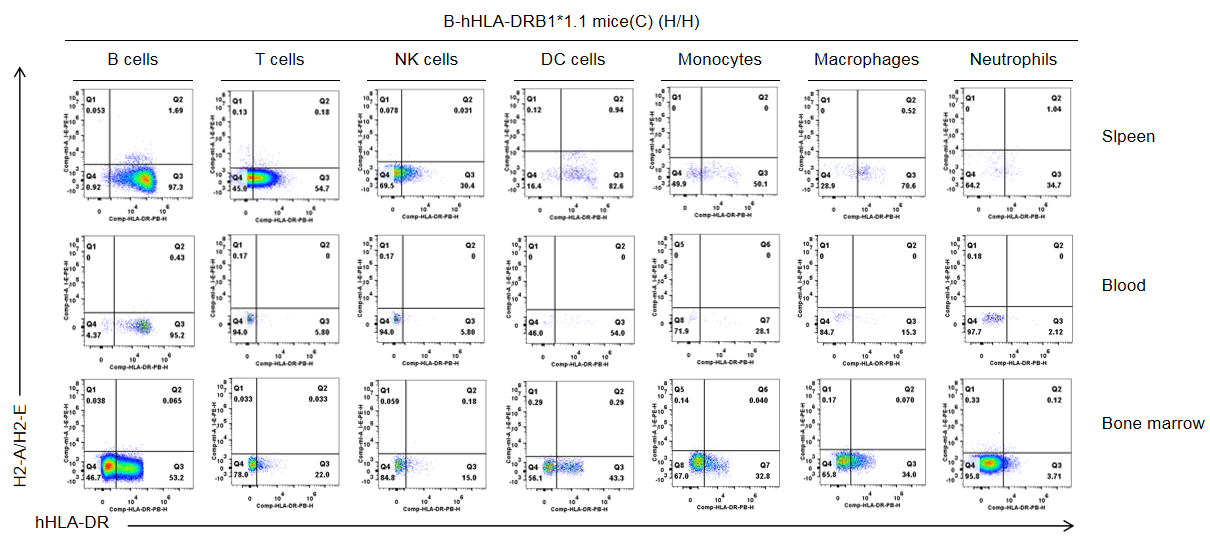
Strain specific HLA-DR expression analysis in homozygous B-hHLA-DRB1*1.1 mice(C) by flow cytometry. Splenocytes, blood cells and bone marrow cells were collected from homozygous B-hHLA-DRB1*1.1 mice (H/H), and analyzed by flow cytometry with species-specific anti-mouse H2-A/H2-E antibody(Biolegend, 107607), and anti-HLA-DR antibody(Biolegend, 307624). HLA-DRB1 was exclusively detectable in immune cells of homozygous B-hHLA-DRB1*1.1 mice(C) but not in the wild-type mice.
Frequency of leukocyte subpopulations in the spleen
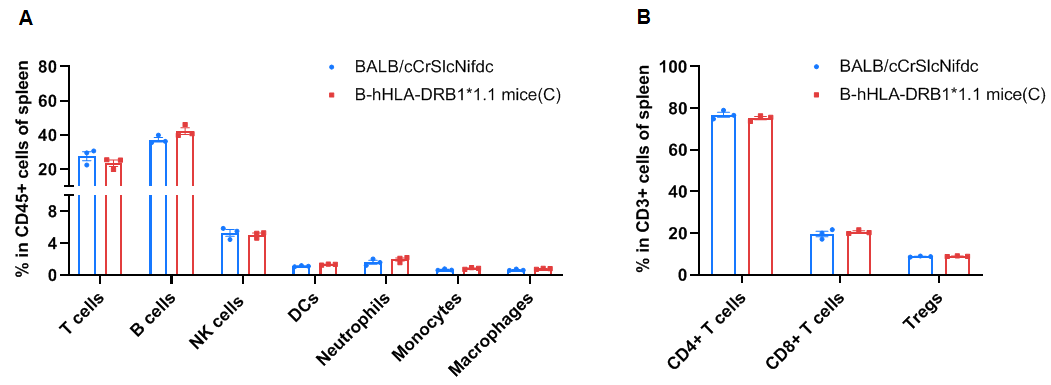
Frequency of leukocyte subpopulations in the spleen by flow cytometry. Splenocytes were isolated from wild-type BALB/cCrSlcNifdc mice (female, n=3, 8-week-old) and homozygous B-hHLA-DRB1*1.1 mice(C) (female, n=3, 8-week-old). A. Flow cytometry analysis of the splenocytes was performed to assess the frequency of leukocyte subpopulations. B. Frequencies of T cell subpopulations. Percentages of T cells, B cells, NK cells, dendritic cells, neutrophils, monocytes, macrophages, CD4+ T cells, CD8+ T cells, and Tregs in B-hHLA-DRB1*1.1 mice(C) were similar to those in BALB/cCrSlcNifdc mice. B cells were increased in homozygous mice. Values are expressed as mean ± SEM. Significance was determined by two-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001.
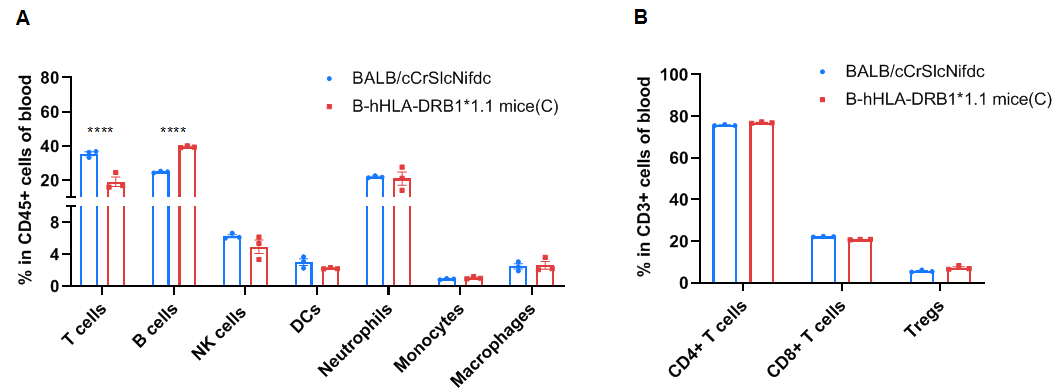
Frequency of leukocyte subpopulations in the blood by flow cytometry. Blood cells were isolated from wild-type BALB/cCrSlcNifdc mice (female, n=3, 8-week-old) and homozygous B-hHLA-DRB1*1.1 mice(C) (female, n=3, 8-week-old). A. Flow cytometry analysis of the blood cells were performed to assess the frequency of leukocyte subpopulations. B. Frequencies of T cell subpopulations. Percentages of T cells, B cells, NK cells, dendritic cells, neutrophils, monocytes, macrophages, CD4+ T cells, CD8+ T cells, and Tregs in B-hHLA-DRB1*1.1 mice(C) were similar to those in BALB/cCrSlcNifdc mice. B cells were increased and T cells were decreased in homozygous mice. Values are expressed as mean ± SEM. Significance was determined by two-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001.
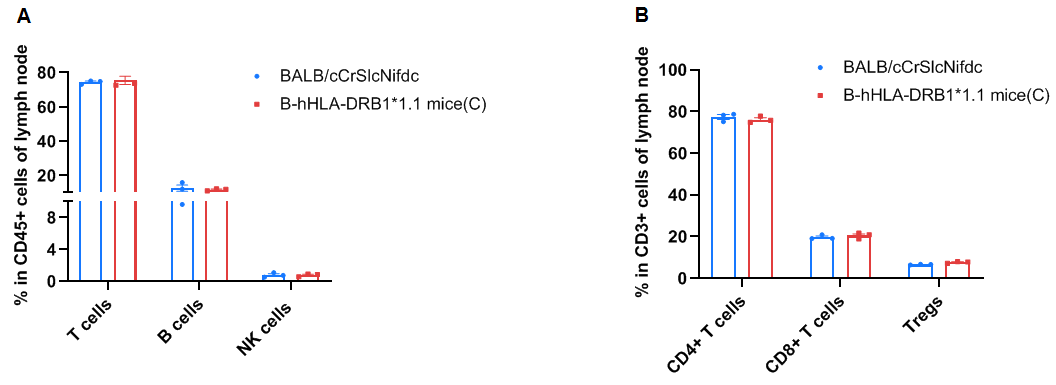
Frequency of leukocyte subpopulations in the lymph node by flow cytometry. The lymph node cells were isolated from wild-type BALB/cCrSlcNifdc mice (female, n=3, 8-week-old) and homozygous B-hHLA-DRB1*1.1 mice(C) (female, n=3, 8-week-old). A. Flow cytometry analysis of the lymph node cells were performed to assess the frequency of leukocyte subpopulations. B. Frequencies of T cell subpopulations. Percentages of T cells, B cells, NK cells, dendritic cells, neutrophils, monocytes, macrophages, CD4+ T cells, CD8+ T cells, and Tregs in B-hHLA-DRB1*1.1 mice(C) were similar to those in BALB/cCrSlcNifdc mice. Values are expressed as mean ± SEM. Significance was determined by two-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001.









